For a complete list of publications, please visit
Google Scholar or ORCID
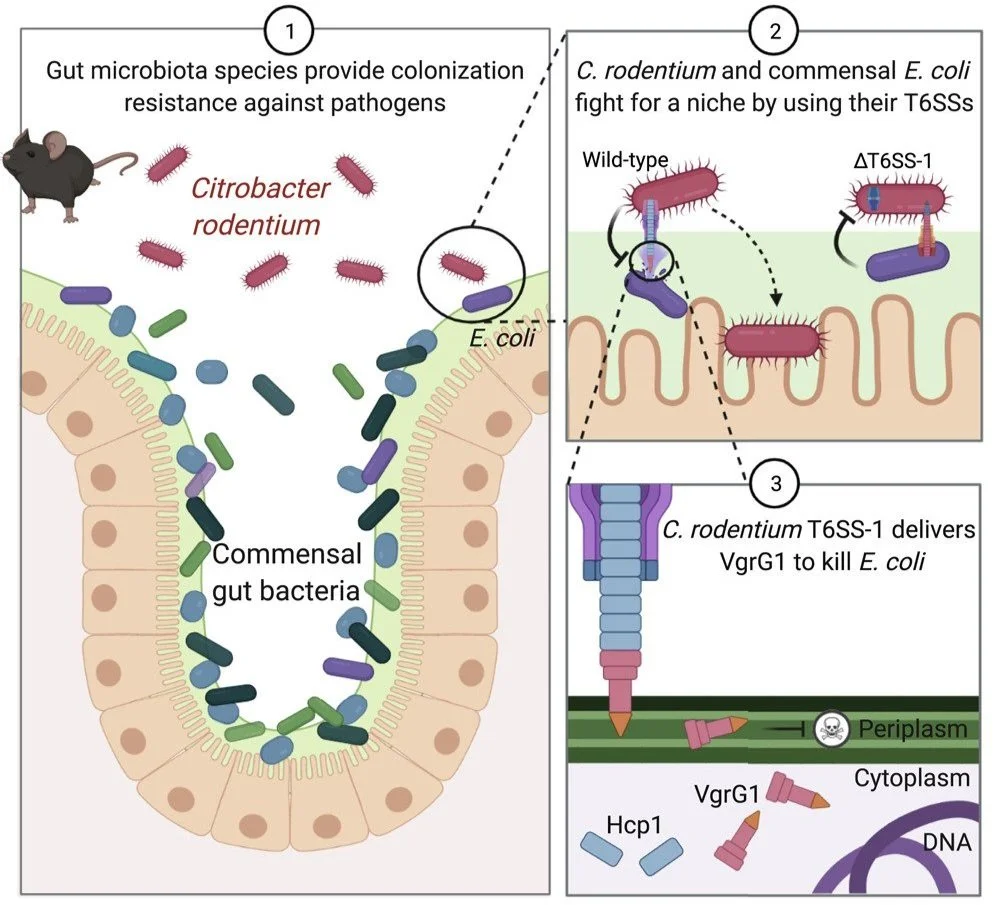
Type VI secretion systems of pathogenic and commensal bacteria mediate niche occupancy in the gut
Serapio-Palacios, A., Woodward, S.E., Vogt, S.L., Deng, W., Creus-Cuadros, A., Huus, K.E., Cirstea, M., Gerrie, M., Barcik, W., Yu, H., Finlay, B.B. (2022). Cell Reports, 39(4).
This study investigates the role of Type VI Secretion System (T6SS) in mediating bacterial competition within the gut microbiota. We demonstrated that both pathogenic and commensal Enterobacteraceae utilize the T6SS to compete for a niche and eliminate competitors. By deploying the T6SS, Citrobacter rodentium can directly target commensal E. coli, iallowing the pathogen to colonize and cause desease
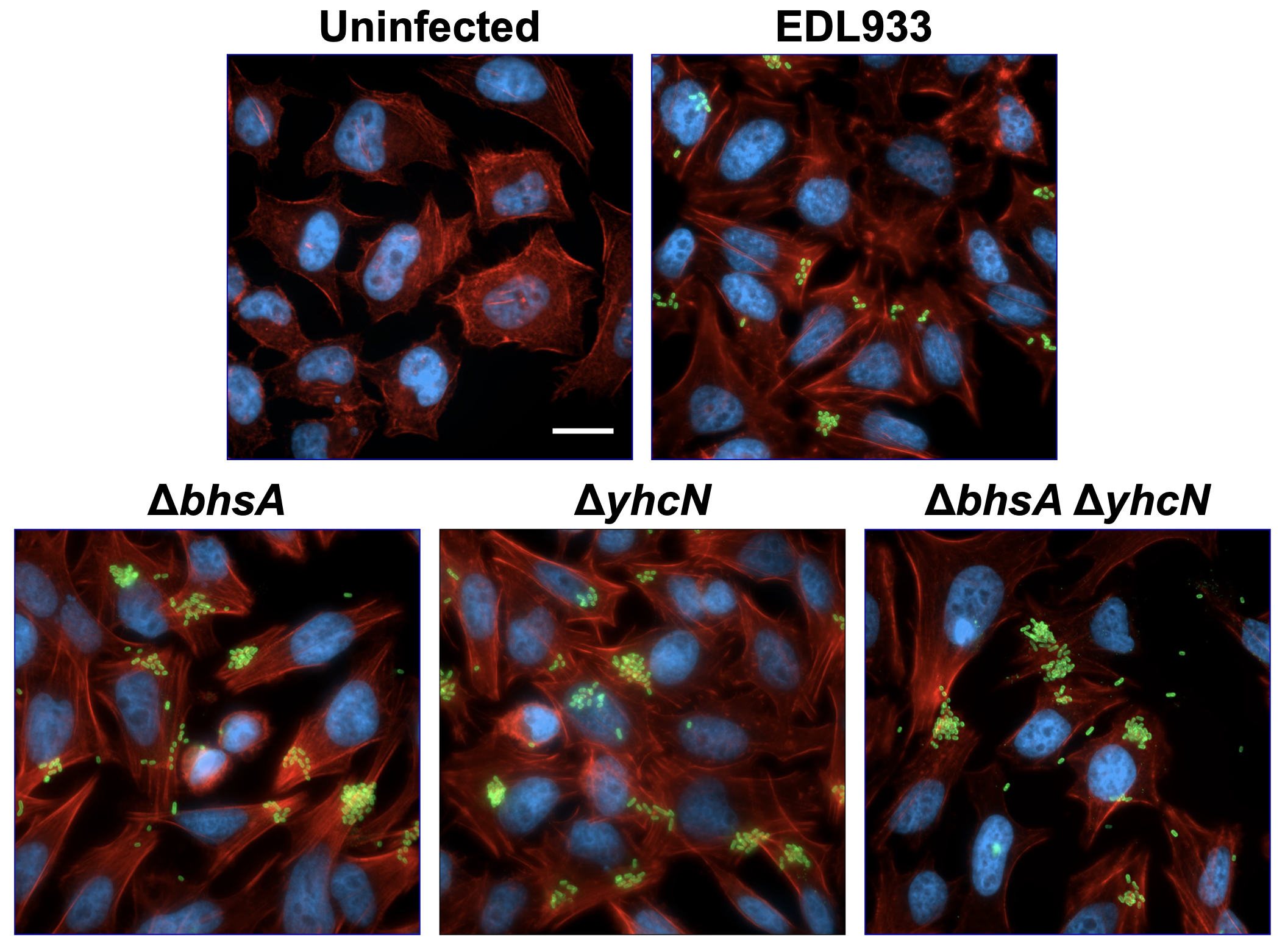
Enterohemorrhagic Escherichia coli responds to gut microbiota metabolites by altering metabolism and activating stress responses.
Vogt, S.L.*, Serapio-Palacios, A.*, Woodward, S.E., Santos, A.S., de Vries, S.P.W., Daigneault, M., Brandmeier L.V., Grant A.J., Maskell D.J., Allen-Vercoe E., Finlay, B.B. (2023) Gut microbes, 15(1), 2190303. *These authors contributed equally to this study.
We investigated how Enterohemorrhagic Escherichia coli (EHEC) adapts to the presence of gut microbiota metabolites. Using RNA-Seq and Tn-Seq analyses, we found that EHEC adjusts its metabolism and activates stress responses when exposed to metabolites produced by a defined human gut microbiota consortium (MET-1). Specifically, EHEC upregulates genes involved in maintaining redox balance and downregulates biotin biosynthesis genes, reflecting the high biotin levels in the environment. Additionally, genes associated with envelope and oxidative stress responses are activated, indicating that EHEC perceives microbiota metabolites as stressors.
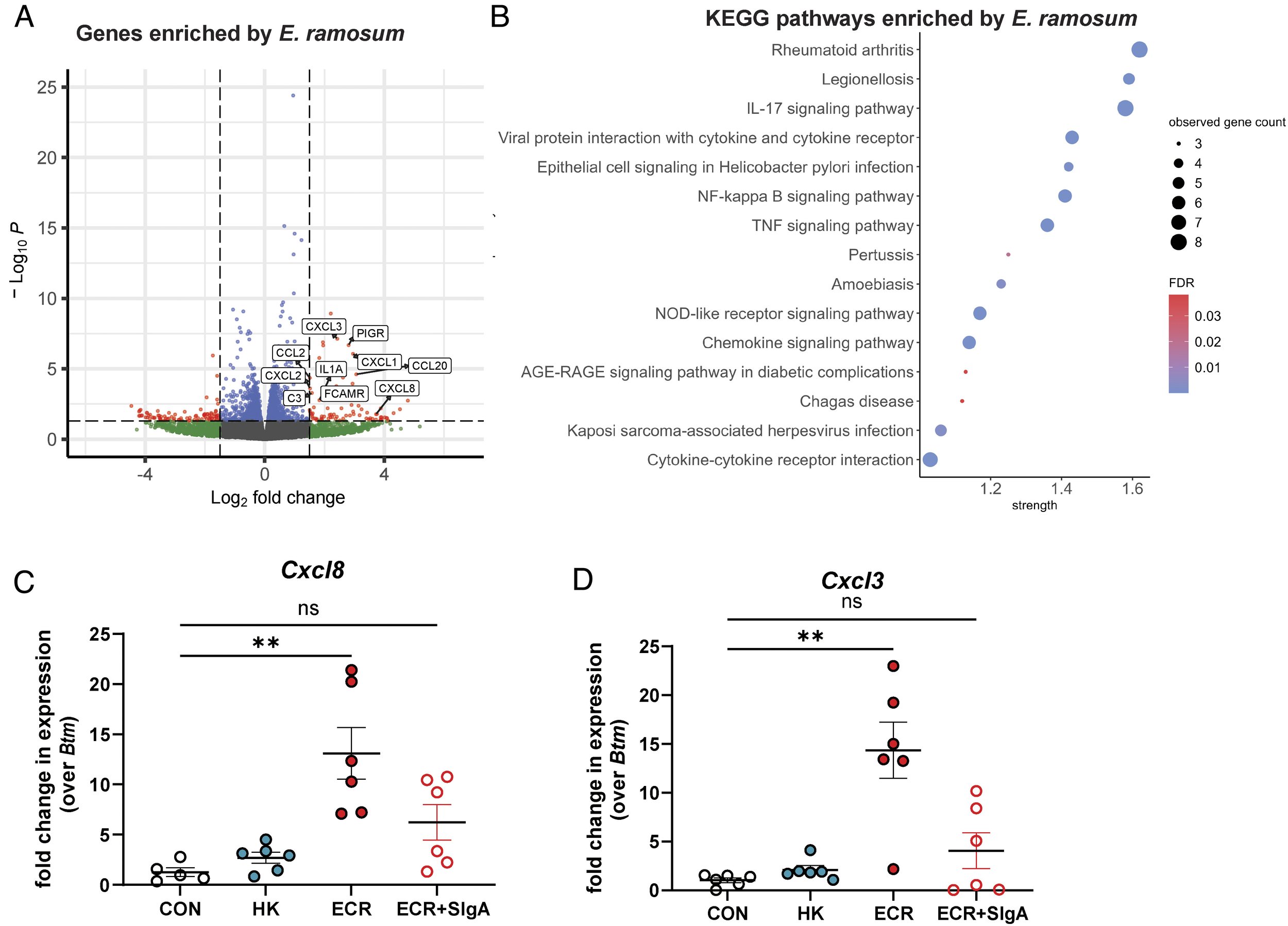
Human milk IgA promotes normal immune development by limiting Th17-inducing Erysipelatoclostridium ramosum in the infant gut
K. Donald, A. Serapio-Palacios, T. Bozorgmehr, M. Ma, M.A.I. Garcia, C. Petersen,P. Mandhane, P. Subbarao, T.J. Moraes, E. Simons, S. Turvey, M.B. Azad, & B.B. Finlay. (2025) Proc. Natl. Acad. Sci. U.S.A. 122 (28) e2501030122.
The infant gut microbiota plays a vital role in immune development. Secretory IgA (SIgA), the most abundant antibody in the gut, is a key modulator of microbiota composition throughout life. Maternal milk is the primary source of intestinal SIgA for the developing infant, but the ability of maternal milk SIgA to guide microbe-mediated immune imprinting remains poorly understood. Here, we characterize the microbiota reactivity of human milk SIgA, demonstrating relationships between maternal SIgA and infant gut microbiota composition. This leads to the identification and characterization of E. ramosum, a microbe that is controlled by milk SIgA and plays an important role in immune development worthy of further consideration in studies of allergy prevention at the maternal–infant interface.
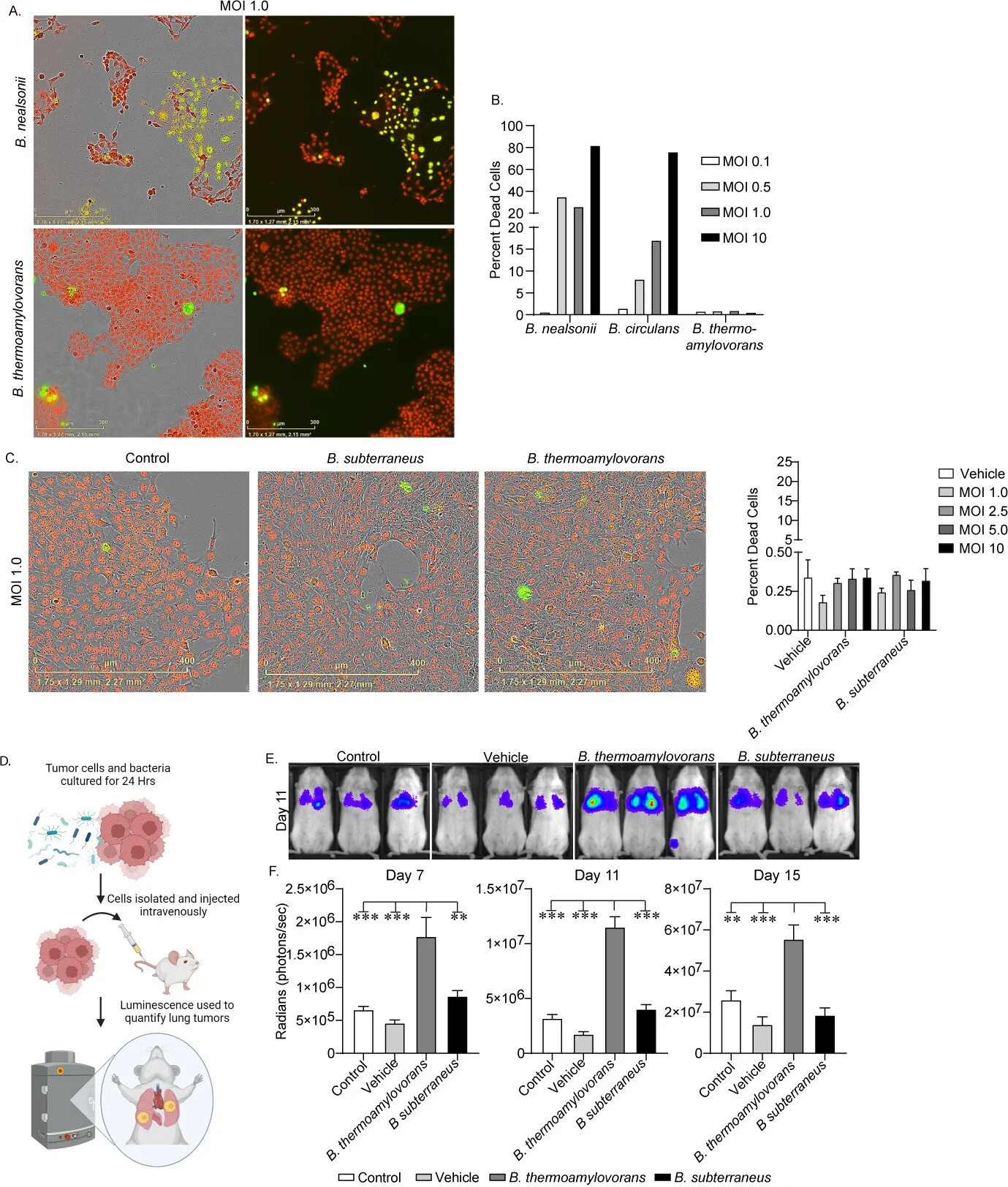
Identification of intratumoral bacteria that enhance breast tumor metastasis
Gerbec ZJ, Serapio-Palacios A, Metcalfe-Roach A, Krekhno Z, Bar-Yoseph H, Woodward SE, Pena-Díaz J, Nemirovsky O, Awrey S, Moreno SH, Beatty S, Kong E, Radisavljevic N, Cirstea M, Chafe S, McDonald PC, Aparicio S, Finlay BB, Dedhar S. (2025)mBio. 12;16(3):e0359524.
We identified specific intratumoral bacteria that promote breast cancer metastasis. Using murine models of metastatic and non-metastatic triple-negative breast cancer, we isolated and characterized bacteria from tumor tissues. One species, Bacillus thermoamylovorans, was found exclusively in metastatic tumors and was shown to increase the metastatic burden in vivo. Through co-culture experiments and metabolomic profiling, this bacterium was demonstrated to reprogram tumor cell metabolism and activate pro-metastatic pathways. This work provides a proof-of-concept that bacterial functional traits, not just taxonomy, can influence cancer progression, paving the way for novel microbiome-informed cancer therapies or diagnostics.
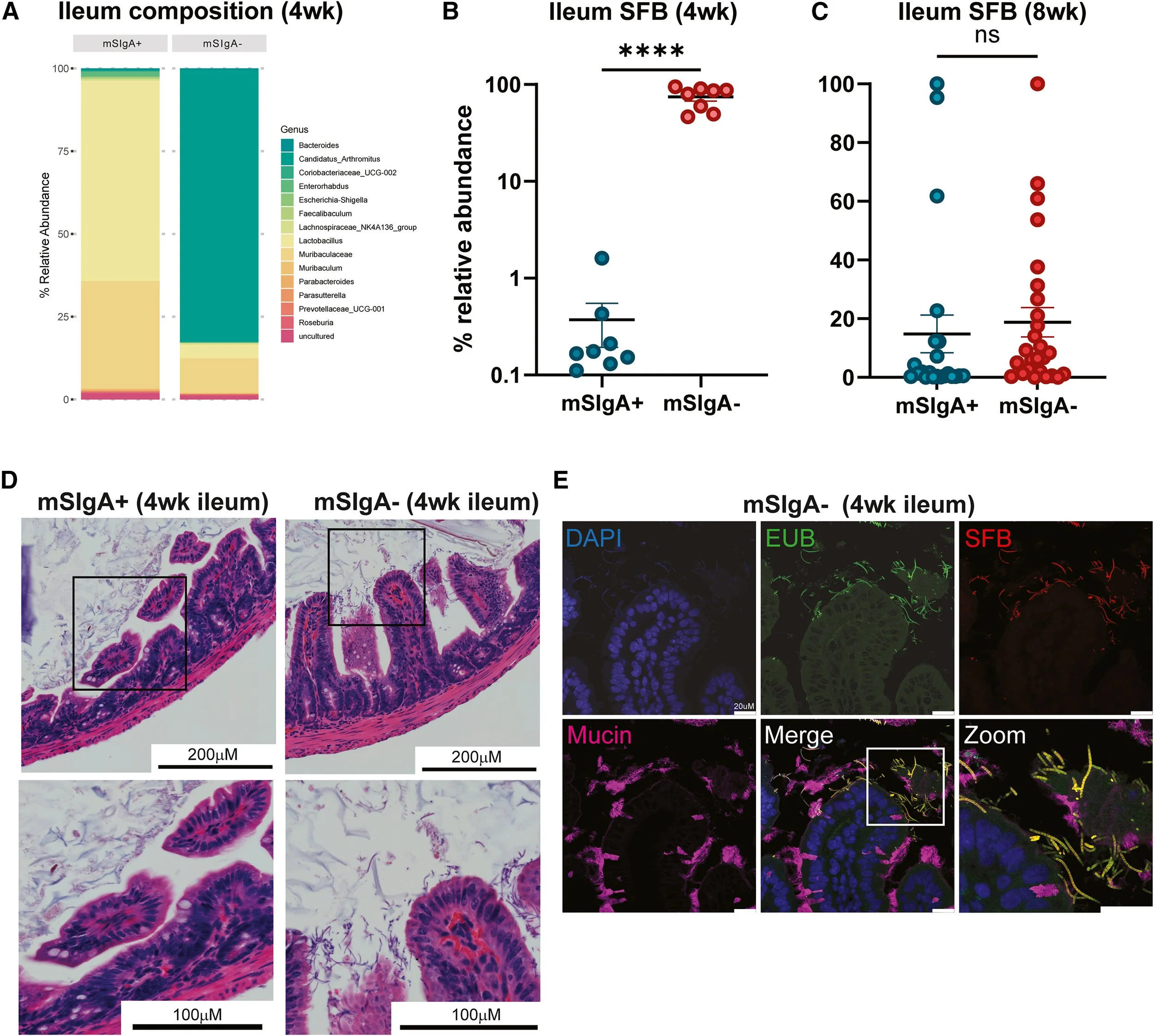
Secretory IgA in Breast Milk Protects against Asthma through Modulation of the Gut Microbiota.
Donald, K., Serapio-Palacios, A., Gerbec, Z.; Bozorgmehr, T., Holani, R., Cruz, A. R., Schnupf, P., Finlay, B. B. (2024) Cell Reports 2024, 43 (10).
We elucidate the protective role of maternal secretory immunoglobulin A (SIgA) in breast milk against the development of asthma in offspring. Using a mouse model, the researchers demonstrate that SIgA limits the colonization of segmented filamentous bacteria (SFB) in the neonatal small intestine. SFB is known to drive premature and elevated Th17 immune responses, which are associated with inflammation. By restricting SFB colonization, SIgA modulates early-life gut microbiota composition, leading to a balanced immune environment that reduces the risk of allergic airway inflammation. These findings highlight the significance of the gut-lung axis in asthma pathogenesis and underscore the importance of breastfeeding in shaping neonatal immunity.