Pathogen-Microbiota-Host Interactions
Molecular mechanism of host-pathogen interactions
We investigate how mammalian host cells respond to enteric infections, with a focus on the strategies used by bacterial pathogens to hijack host pathways and promote disease. Our research centers on secretion systems — molecular nanomachines that enable pathogens to deliver “effector” proteins directly into host cells. These effectors reprogram host cellular functions to allow bacterial pathogens to colonize and evade the immune system. We aim to identify the effector repertoires of enteric pathogens and uncover how these proteins manipulate host responses at the cellular and molecular level. To do so, we develop and integrate in vivo animal models, in vitro infection systems, and advanced genetic tools.
Interbacterial competition in the gut
We investigate how pathogens and commensal bacteria interact and compete within the gut microbiome. A key mechanism driving these interactions is the Type VI Secretion System (T6SS)—a contact-dependent molecular weapon that allows bacteria to inject toxic effector proteins into neighboring cells. Through this system, bacteria can kill competitors, reshaping the gut microbial composition and influencing host health. Our research centers on Escherichia coli and other members of the Enterobacteriaceae family, examining how they deploy the T6SS to establish dominance in the intestinal environment. To uncover these dynamics, we employ multiple approaches, including in vitro bacterial competition assays, high-resolution microscopy, next-generation sequencing, and in vivo animal models. Together, these tools allow us to visualize microbial warfare and unravel the molecular strategies that shape microbial communities.
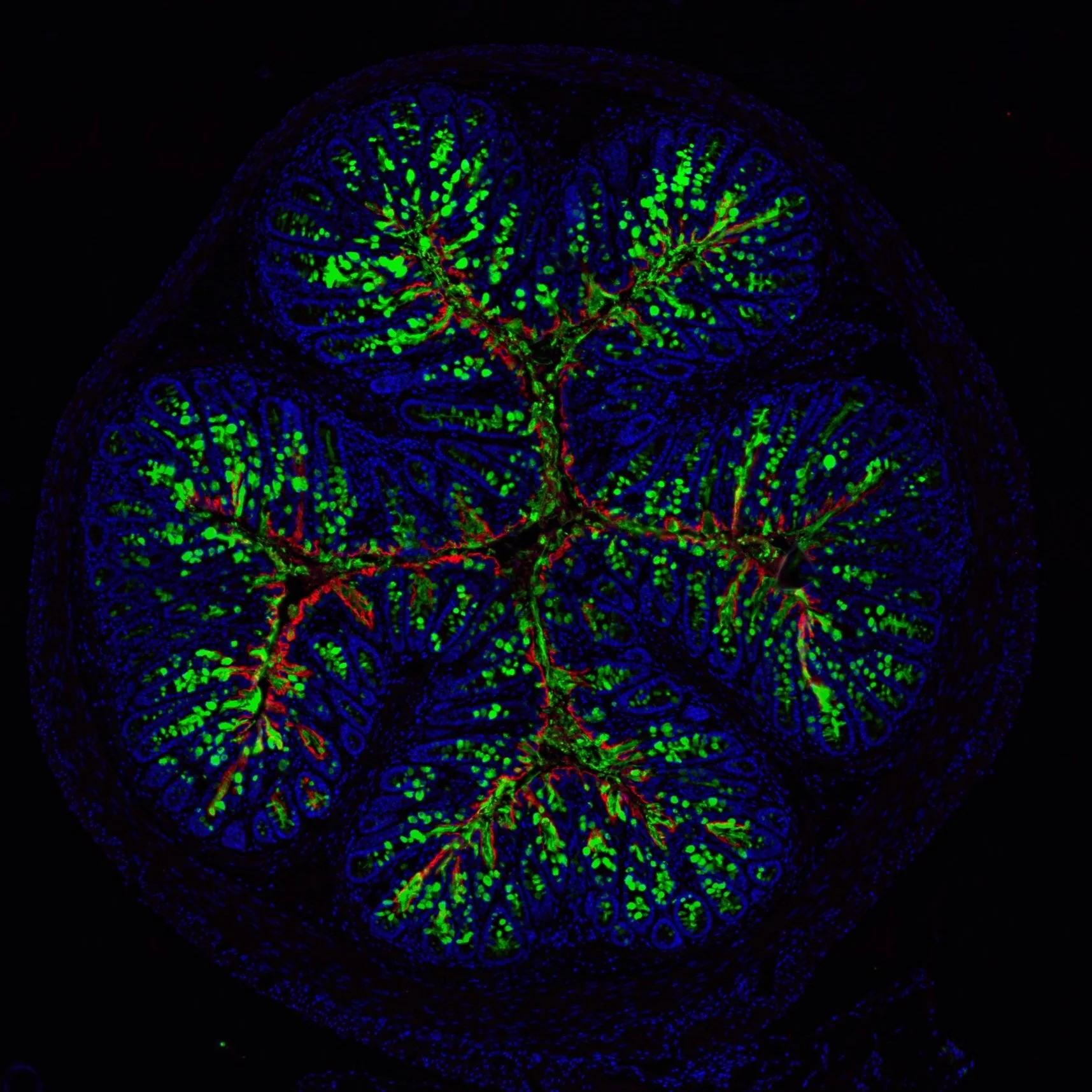
Microbial factors involved in inflammation and disease
Our lab investigates how microbes, their associated proteins and metabolites contribute to inflammation and disease. While many members of the gut microbiome are beneficial, certain bacterial species possess virulence traits that can disrupt host homeostasis. We focus on understanding how bacterial effectors, toxins, and surface molecules trigger inflammatory pathways, compromise the epithelial barriers, and modulate immune responses. By dissecting these host-microbe interactions at the molecular level, we aim to identify the bacterial determinants that can tip the balance from tolerance to inflammation, providing crucial insights into the causes of chronic inflammatory diseases and microbiome-mediated disorders.